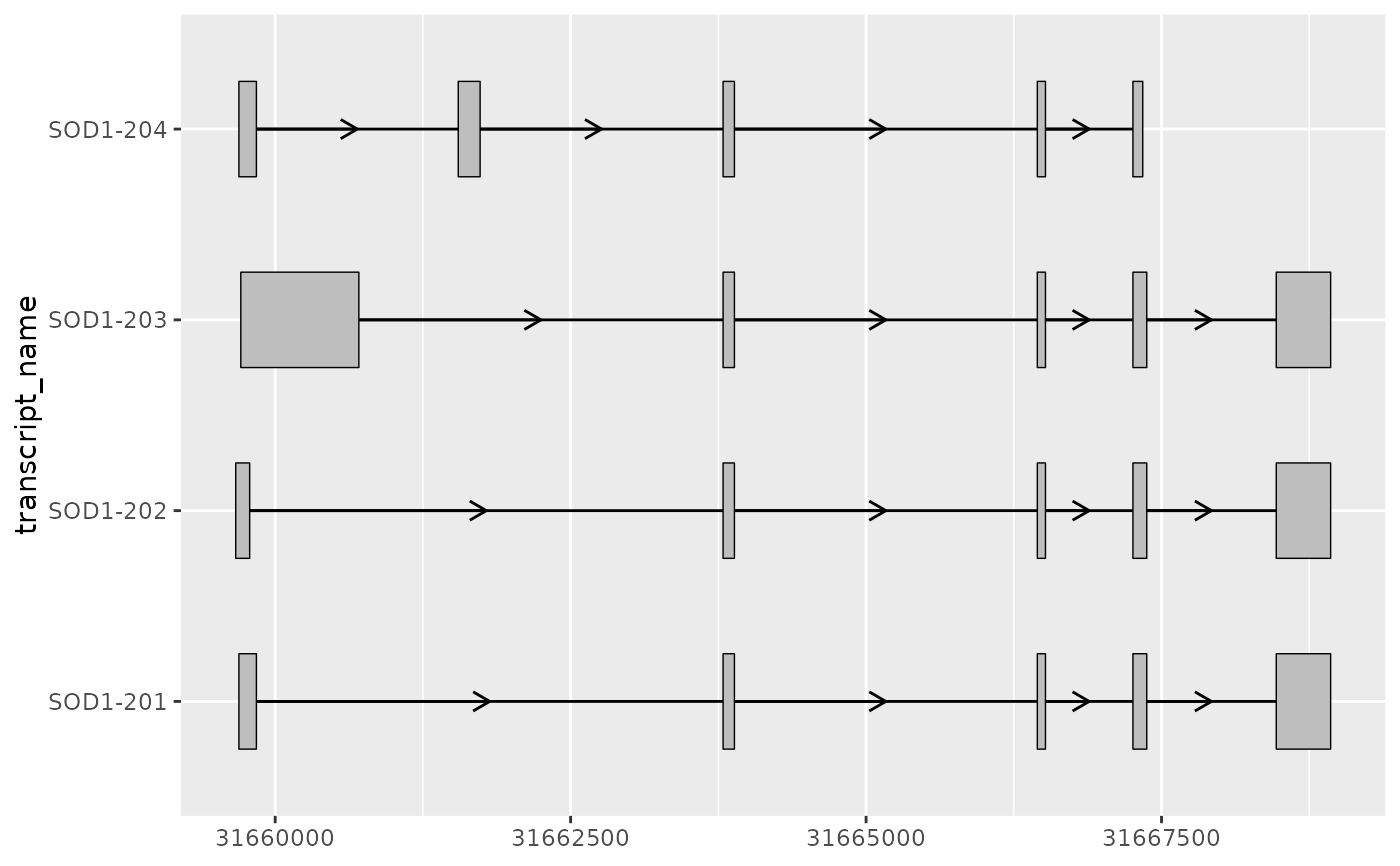
Given a set of exons, to_intron() will return the corresponding introns.
Arguments
- exons
data.frame()contains exons which can originate from multiple transcripts differentiated bygroup_var.- group_var
character()if input data originates from more than 1 transcript,group_varmust specify the column that differentiates transcripts (e.g. "transcript_id").
Value
data.frame() contains the intron co-ordinates.
Details
It is important to note that, for visualization purposes, to_intron()
defines introns precisely as the exon boundaries, rather than the intron
start/end being (exon end + 1)/(exon start - 1).
Examples
library(magrittr)
library(ggplot2)
# to illustrate the package's functionality
# ggtranscript includes example transcript annotation
sod1_annotation %>% head()
#> # A tibble: 6 × 8
#> seqnames start end strand type gene_name transcript_name
#> <fct> <int> <int> <fct> <fct> <chr> <chr>
#> 1 21 31659666 31668931 + gene SOD1 NA
#> 2 21 31659666 31668931 + transcript SOD1 SOD1-202
#> 3 21 31659666 31659784 + exon SOD1 SOD1-202
#> 4 21 31659770 31659784 + CDS SOD1 SOD1-202
#> 5 21 31659770 31659772 + start_codon SOD1 SOD1-202
#> 6 21 31663790 31663886 + exon SOD1 SOD1-202
#> # ℹ 1 more variable: transcript_biotype <chr>
# extract exons
sod1_exons <- sod1_annotation %>% dplyr::filter(type == "exon")
sod1_exons %>% head()
#> # A tibble: 6 × 8
#> seqnames start end strand type gene_name transcript_name
#> <fct> <int> <int> <fct> <fct> <chr> <chr>
#> 1 21 31659666 31659784 + exon SOD1 SOD1-202
#> 2 21 31663790 31663886 + exon SOD1 SOD1-202
#> 3 21 31666449 31666518 + exon SOD1 SOD1-202
#> 4 21 31667258 31667375 + exon SOD1 SOD1-202
#> 5 21 31668471 31668931 + exon SOD1 SOD1-202
#> 6 21 31659693 31659841 + exon SOD1 SOD1-204
#> # ℹ 1 more variable: transcript_biotype <chr>
# to_intron() is a helper function included in ggtranscript
# which is useful for converting exon co-ordinates to introns
sod1_introns <- sod1_exons %>% to_intron(group_var = "transcript_name")
sod1_introns %>% head()
#> # A tibble: 6 × 8
#> seqnames strand type gene_name transcript_name transcript_biotype start
#> <fct> <fct> <chr> <chr> <chr> <chr> <int>
#> 1 21 + intron SOD1 SOD1-204 processed_transcript 31659841
#> 2 21 + intron SOD1 SOD1-202 protein_coding 31659784
#> 3 21 + intron SOD1 SOD1-204 processed_transcript 31661734
#> 4 21 + intron SOD1 SOD1-201 protein_coding 31659841
#> 5 21 + intron SOD1 SOD1-203 processed_transcript 31660708
#> 6 21 + intron SOD1 SOD1-202 protein_coding 31663886
#> # ℹ 1 more variable: end <int>
# this can be particular useful when combined with
# geom_range() and geom_intron()
# to visualize the core components of transcript annotation
sod1_exons %>%
ggplot(aes(
xstart = start,
xend = end,
y = transcript_name
)) +
geom_range() +
geom_intron(
data = to_intron(sod1_exons, "transcript_name")
)