Note: install the development version of biocthis via
devtools::install_github("lcolladotor/biocthis")and any additional dependencies.
Outline
- Why make R/Bioconductor packages?
- What do R packages consist of?
- Testing fundamentals.
- Package development packages -
usethis,devtoolsandbiocthis. - Package development workflow.
- Additional resources.
- Making your own Bioconductor-friendly package.
Why make R/Bioconductor packages?
R packages
- Share functions with others/yourself.
- Improving the organization of your functions - e.g. mentality, documentation, continuous updates.
- Integrated unit testing to keep your functions robust to future changes.
Useful resources:
- R packages book
- Your first R package in 1 hour
Bioconductor packages
- Share code more widely with Bioconductor community.
- Code review, learning about developing software - forced compliance with Bioconductor standards.
- More citations
- CV/reputation points.
What do R packages consist of?
States of R packages
- The states of R packages is explained well here.
- 5 states:
- source - A set files/directories in a set structure. The state you develop with and encounter when cloning a GitHub repository.
- bundles - Source compressed into a single tar.gz (e.g. through
devtools::build()) - binary - Binary version (e.g. distributed by CRAN). Can be generated through
devtools::build(binary = TRUE). - installed - Either the binary or bundle that’s decompressed and placed into your R package library (using
install.packages()) - in-memory - Load a package into memory using
library()ordevtools::load_all().
Structure of R packages
R packages have several key components, which are shown in screen-shot taken from the biocthis GitHub repository below:
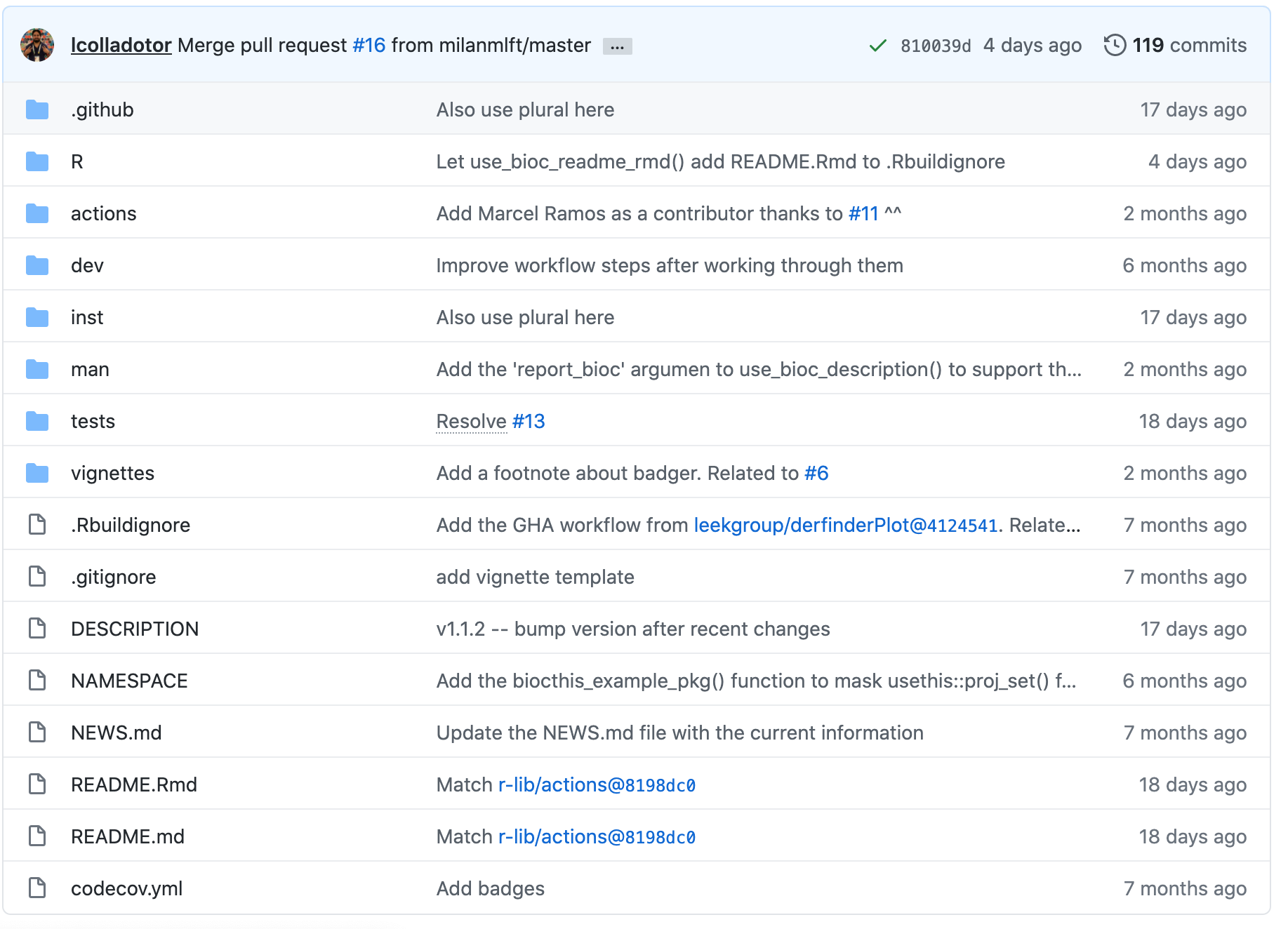
- .github - GitHub actions workflow.
- R - Contains all the code for the functions of your package.
- dev - Package development helper scripts from
biocthis. - inst - Additional raw data you would like available to the user.
- man - Function documentation generated by Roxygen.
- tests - Unit tests for functions.
- vignettes - Vignette description usage of your package
- DESCRIPTION - Author details, license, dependencies and general summary of your package.
- README.md - Documentation shown at the GitHub repository.
- NEWs.md - Collection of updates for package for users to scan through.
Testing fundamentals
3 levels of testing:
- Unit testing individual functions.
- R CMD Check or BiocCheck checks that you’re package can be built, installed and complies to R/Bioconductor package guidelines.
- Testing across multiple operating systems using GHA.
Unit testing
- For complicated code, ensures future updates do not break existing functionality.
- You do this already, just manually.
testhatautomates this for R packages.- Rule of thumb: If you catch a bug, write a test.
- Metrics, try to be fast + comprehensive.
R CMD Check or BiocCheck
R CMD Checkthat your package meets certain criteria:- Runs all unit tests
- Have you set all the dependencies you need?
- Have you documented your functions?
- Have you written a proper
DESCRIPTION? - Have you used
:::? - Lots more… see R packages page on R CMD Check
BiocCheckis more stringent:- Are your functions less than 80 lines?
- Do >80% have examples?
- Reports
ERROR/WARNING/NOTEs.
Testing across Linux/Mac/Windows
- A great lecture on GHA basics from Jim Hester can be found here.
- GitHub actions is used in this case for testing, but is very flexible.
- On an event, do something.
- Written in yet-another-markdown-language (YAML).
- GitHub has “runners” based on Linux/Mac/Windows OS (and permits dockers).
- We run
R CMD CheckandBiocCheckon all 3 to check for OS-specific issues.
Package development packages
usethis, devtools and biocthis are convenient helper packages for developing R/bioconductor packages respectively.
usethis
- usethis has a lot of functions to enable easy package development.
usethis::create_package()- creates package skeleton.usethis::use_r()- creates .R function skeleton.usethis::use_test()- create test skeleton.usethis::use_package()- add a dependency toImports.usethis::use_git()- connects current package to git.usethis::use_github()- create a GitHub repo for current repo.usethis::use_github_action()- creates GHA skeleton.
devtools
- devtools is aimed at “making your life as a package developer easier”
devtools::load_all()- load your current packagedevtools::test()- run all unit testsdevtools::check()- runR CMD Checkdevtools::document()- document with roxygen2devtools::build()- build your current packagedevtools::install()- installs your current package
biocthis
- biocthis extends
usethisspecifically for Bioconductor packages:- Templates for creating
DESCRIPTION,README.mdand vignettes - Styles using
BiocStyle - Sets up a GitHub action workflow on all 3 OS (including using the Bioconductor docker for Linux) that includes
R CMD CheckandBiocCheck. - Deploys a
pkgdownpage for your package. - Sets up code coverage check.
- Templates for creating
- Still under active development.
Package development workflow
A top-level workflow for modifying your package, then running tests and updating via git is shown below.

Additional resources
- What makes good tests/documentation? Where should I put raw vs processed vs internal data? What else does
R CMD Checkcheck? The R packages book is highly recommended. - Writing
Rfunctions - good practices can be found in the tidyverse design guide. testthat- 3rd edition- How to write your own GHA workflow? A good place to start with using GHA in R can be found here.
- Submission/maintenance of Bioconductor packages. Info regarding submissions can be found here and maintaining packages here.
- An alternative making Bioconductor packages workshop can be found here. This covers in detail additional topics such as submitting your package to Bioconductor.
Making your own Bioconductor-friendly package
- Create an R package skeleton with
usethis::create_package("path_to_package"). - Create
dev/folder withbiocthisscripts usingbiocthis::use_bioc_pkg_templates(). - Run
available::available("package_name")to check package name availability. - Run through
dev/02_git_github_setup.R(adding GitHub PAT and ssh keys if not set up already). Check your initial skeleton has been pushed to GitHub repo. - Finishing setting up
dev/03_core_files.R. You may need to installknitcitationsfrom GitHub withremotes::install_github("cboettig/knitcitations"). You may also need to install your package viadevtools::install(".")before runningpkgdown::deploy_to_branch(). usethis::use_r("function_name")- add your own function with documentation and dependencies viausethis::use_package("package_name"): example function get_sum_stats.R.usethis::use_test("function_name")- add testing for your function: example tests test-get_sum_stats.R.- Run unit tests via
devtools::test(). - Run
04_update.Rto auto-style and document your code. - Run R CMD Check via
devtools::check(). - If passed then push your changes to GitHub. Review
ERROR/WARNING/NOTESthen make any necessary changes (e.g. morebiocViews) and update.
Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.0.3 (2020-10-10)
## os macOS Big Sur 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_GB.UTF-8
## ctype en_GB.UTF-8
## tz Europe/London
## date 2020-12-15
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date lib source
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.2)
## backports 1.2.0 2020-11-02 [1] CRAN (R 4.0.2)
## broom 0.7.2 2020-10-20 [1] CRAN (R 4.0.2)
## cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.0.2)
## cli 2.2.0 2020-11-20 [1] CRAN (R 4.0.2)
## colorspace 2.0-0 2020-11-11 [1] CRAN (R 4.0.2)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 4.0.2)
## DBI 1.1.0 2019-12-15 [1] CRAN (R 4.0.2)
## dbplyr 2.0.0 2020-11-03 [1] CRAN (R 4.0.2)
## digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.2)
## dplyr * 1.0.2 2020-08-18 [1] CRAN (R 4.0.2)
## ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.2)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.1)
## fansi 0.4.1 2020-01-08 [1] CRAN (R 4.0.2)
## forcats * 0.5.0 2020-03-01 [1] CRAN (R 4.0.2)
## fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
## generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
## ggplot2 * 3.3.2 2020-06-19 [1] CRAN (R 4.0.2)
## glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.2)
## haven 2.3.1 2020-06-01 [1] CRAN (R 4.0.2)
## hms 0.5.3 2020-01-08 [1] CRAN (R 4.0.2)
## htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.2)
## httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
## jsonlite 1.7.1 2020-09-07 [1] CRAN (R 4.0.2)
## knitr * 1.30 2020-09-22 [1] CRAN (R 4.0.2)
## lifecycle 0.2.0 2020-03-06 [1] CRAN (R 4.0.2)
## lubridate 1.7.9.2 2020-11-13 [1] CRAN (R 4.0.2)
## magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.2)
## modelr 0.1.8 2020-05-19 [1] CRAN (R 4.0.2)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.2)
## pillar 1.4.7 2020-11-20 [1] CRAN (R 4.0.2)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.2)
## purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.2)
## R6 2.5.0 2020-10-28 [1] CRAN (R 4.0.2)
## Rcpp 1.0.5 2020-07-06 [1] CRAN (R 4.0.2)
## readr * 1.4.0 2020-10-05 [1] CRAN (R 4.0.2)
## readxl 1.3.1 2019-03-13 [1] CRAN (R 4.0.2)
## reprex 0.3.0 2019-05-16 [1] CRAN (R 4.0.2)
## rlang 0.4.9 2020-11-26 [1] CRAN (R 4.0.2)
## rmarkdown 2.5 2020-10-21 [1] CRAN (R 4.0.3)
## rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.0.2)
## rvest 0.3.6 2020-07-25 [1] CRAN (R 4.0.2)
## scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.2)
## sessioninfo * 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
## stringi 1.5.3 2020-09-09 [1] CRAN (R 4.0.2)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.2)
## tibble * 3.0.4 2020-10-12 [1] CRAN (R 4.0.2)
## tidyr * 1.1.2 2020-08-27 [1] CRAN (R 4.0.2)
## tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.2)
## tidyverse * 1.3.0 2019-11-21 [1] CRAN (R 4.0.2)
## vctrs 0.3.5 2020-11-17 [1] CRAN (R 4.0.2)
## withr 2.3.0 2020-09-22 [1] CRAN (R 4.0.2)
## xfun 0.19 2020-10-30 [1] CRAN (R 4.0.2)
## xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.2)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.2)
##
## [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library